Curation Policy
Microbial Signature Curation Project Policy[edit | edit source]
Scope of BugSigDB[edit | edit source]
BugSigDB is a comprehensive database of microbial signatures which enables comparison of microbiome studies to previously published results. In general, the scope of papers that BugSigDB is intended to represent includes:
- microbiomes associated with any host (if your host of interest is not yet listed here, ask an administrator to add it)
- differential abundance between different conditions (for example experiment vs control, case vs control, time 1 vs time 2, high exposure vs low exposure). There are rare exceptions to this rule, such as the Body site-typical microbiome signatures for human children and adults.
- both peer-reviewed results and pre-prints are allowed.
Curation workflow[edit | edit source]
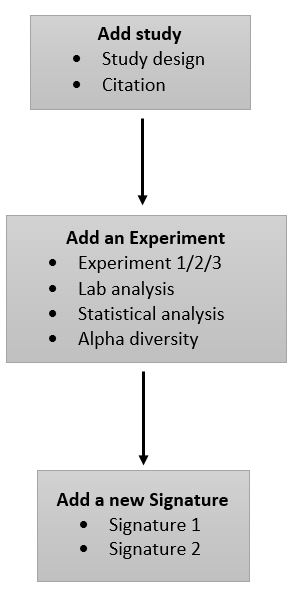
The basic workflow for curation of a new study is as follows:
Add a new study[edit | edit source]
- Add a Study. If possible, enter the PubMed ID (PMID). BSDB will automatically import the rest of the study information based on the PubMed ID. You do not need to enter the DOI, URI, or author information if you enter the PubMed ID. This is the preferred method. If the PubMed ID is not available, you can manually enter the study information.
- Enter the study design. See below for a guide to study designs.
Study Design[edit | edit source]
| Design | Experimental or Observational | Characteristics | Example |
|---|---|---|---|
| Case-Control | Observational | Participants are selected for study based on whether they have (cases) or do not have (controls) the exposure or health outcome of interest. A subset of a larger prospective cohort study or cross-sectional observational study, where participants are selected based on an exposure or health outcome of interest, becomes a "nested" case-control study (Cigarette smoking and the oral microbiome in a large study of American adults). | A comparison of the oral microbiomes of heavy smokers to non-smokers |
| Cross-sectional observational, not case-control | Observational | Cross-sectional observational refers to comparison of different individuals at a point in time, based on observation of their exposure or health outcome of interest. Unlike a case-control, participants are not selected based on this exposure or health outcome - such as an analysis of pack-years of lifetime cigarette exposure that includes participants regardless of smoking history and current smoking status. Individuals can also be compared based on some socio-demographic characteristics (race, educational level, towns, hospitals, etc.) - such as a population-based New York study where the oral microbiomes of different individuals were compared based on various sociodemographic characteristics such as race/ethnicity, family income, education, sex. | Sociodemographic variation in the oral microbiome |
| Randomized Controlled Trial (RCT) | Experimental | An RCT is a study where participants are randomized to receive a treatment (generally a drug, therapy, diet, etc.) The defining characteristic is that the treatment or exposure is assigned intentionally and randomly to participants by the researchers, not that the study occurs in a clinic. | Habitual dietary fibre intake influences gut microbiota response to an inulin-type fructan prebiotic: a randomised, double-blind, placebo-controlled, cross-over, human intervention study |
| Laboratory Experiment | Experimental | This type of study is often done using animal models, such as rats, pigs, mice, etc. The defining feature is that the researcher controls the contrast of interest, rather than just observing it, under controlled laboratory conditions. | Effects of smoking on the lower respiratory tract microbiome in mice |
| Time series / longitudinal observational | Observational | The defining characteristic of this study design is an analysis comparing repeated observations of the same participant over time. This can be before and after an event or at multiple times. This analysis could be performed within a prospective cohort study, but the analysis over time or before and after event attribute distinguishes this from the Prospective Cohort design. | A comparison of the infant gut microbiome before versus after the start of the covid-19 pandemic |
| Prospective Cohort | Observational | In prospective cohort studies, participants are recruited based on a common characteristic (biomarkers, residence, clinical variables, etc.) and they're followed up over time. Over time, some participants in the cohort develop the condition of interest (cases) and some participants don't (controls). | Respiratory Tract Dysbiosis Is Associated with Worse Outcomes in Mechanically Ventilated Patients |
| Meta-analysis | Either | Meta-analysis attempts to pool information from all available, relevant literature or data to inform its conclusions. Pooling of e.g. two datasets or subsets collected by the authors doesn't count. | Pooling of metagenomic studies of Colorectal Cancer to identify biomarkers |
Create an Experiment[edit | edit source]
- An experiment is a comparison between two (or more groups) where they conducted some form of differential abundance analysis on microbial taxa. Most studies conduct multiple comparisons and therefore have multiple experiments.
- For each comparison, create an experiment. Using the "Duplicate this experiment" allows you to easily create a new experiment with the same experimental information. This is useful for creating a new experiment when it only differs slightly from a previous one.
Location of Subjects[edit | edit source]
- This is the country from which study subjects were recruited.
- If the study is an animal or environmental study, curate the country where the samples were collected.
- The country or location is generally stated in the methods section or results section. You may need to google the city, hospital, or health center mentioned to determine the country.
- Can be multiple countries in some studies.
- If no location is mentioned in the article, look at the affiliation of the first and last authors of the paper for their location.
Host Species[edit | edit source]
- Species from which microbiome was sampled (if applicable). Leave blank for environmental studies.
- Drop down options (for the complete and up-to-date list, see Property:Host_species)
- Anolis carolinensis (green anole)
- Anopheles gambiae (African mosquito)
- Arabidopsis thaliana (thale cress)
- Bos taurus (cow)
- Caenorhabditis elegans (roundworm)
- Canis lupus familiaris (dog)
- Danio rerio (zebrafish)
- Drosophilia melanogaster (fruit fly)
- Equus caballus (horse)
- Felis catus (cat)
- Gallus gallus (junglefowl)
- Homo sapiens (human)
- Macaca mulatta (rhesus macaque)
- Monodelphis domestica (opossum)
- Mus musculus (mouse)
- Ornithorhynchus anatinus (platypus)
- Pan troglodytes (chimpanzee)
- Rana pipiens (leopard frog)
- Rattus norvegicus (rat)
- Sus scrofa domesticus (pig)
- Xenopus laevis (African clawed frog)
- Xenopus tropicalis (Western clawed frog)
- Ailuropoda melanoleuca (giant panda)
- Salmo salar (Atlantic salmon)
- Agyrosomus regius (salmon-bass)
Body site[edit | edit source]
- Check environmental ontology through this link for properly reporting the body sites.
- Sometimes body sites can be difficult to find in the ontology. Remember that some body sites have multiple names (e.g. "mouth" and "oral cavity"). If you are having trouble figuring out the body site, please ask on the Slack.
- If the drop down doesn’t contain the condition, please use the link to find appropriate ontology.
Condition[edit | edit source]
- Condition is the disease, medical treatment, drug, health status variable, genetic variant, or environmental factor that the study contrasts on.
- The condition contrasted on is almost never "microbiome measurement" or similar conditions that refers to measurement of the microbiome.
- Examples of conditions include: type 1 diabetes, kidney transplant, depression, colorectal cancer, exposure to air pollution, smoking status measurement
- Check experimental ontology through the EFO for properly reporting the conditions.
- If the drop down doesn’t contain the condition, please use the link to find appropriate ontology.
Contrast[edit | edit source]
- The contrast between two groups (unexposed vs. exposed)
- Group 0 name: Corresponds to the control (unexposed) group for case-control and other studies
- Group 1 name: Corresponds to the case (exposed) group for case-control studies
- In some cases, there are groups with low vs. high severity of a disease, use group 0 for low severity and group 1 for high severity cases.
- Don’t switch the sample size of exposed groups (cases) with unexposed groups (controls).
- Sample sizes should be the final analytic sample size used for the experiment. Sometimes the paper will report the number initially screened or note that some samples were excluded due to quality control issues.
Exclusion Criteria[edit | edit source]
- Antibiotics only.
- We do not need to curate any other exclusion criteria for any other medicines, treatments, or conditions.
- Include the time frame given for antibiotic exclusion e.g. 2 months, 3 weeks etc.
- The following formatting is preferred for easier analysis: X <units of time> (e.g. "2 months", "90 days", or "6 weeks")
- Please do not include any exclusion criteria about anything other than antibiotics.
Lab Analysis[edit | edit source]
- Add Sequencing type
- Add Sequencing platform
- This information can be found in the methods section.
Sequencing type[edit | edit source]
- Two main types: 16S and WMS (Whole Metagenome Sequencing)
- 16S variable regions (lower and upper) e.g. V3-V4 (V3 will be lower bound and V4 will be upper bound).
- If there is only one variable region given e.g V4, use only lower bound.
- Rarely, the paper may be missing the sequencing variable region for 16S. If so, leave the variable region blank.
Sequencing Platform[edit | edit source]
- Manufacturer and experimental platform used for quantifying microbial abundance.
- Use the drop down options to select the appropriate platform.
Statistical Analysis[edit | edit source]
Data Transformations[edit | edit source]
Raw Counts (Deseq2, negative binomial/poisson regression)
# of times that taxon was observed in the sample
Relative Abundance (t-test, Mann-Whitney's U, linear regression, etc.) *should be basic assumption*
Compared to the overall composition, what % of the microbiome was each taxon?
Centered log ratio (CORNCOB, linear regression, ANCOM but not always)
Take the log of the relative abundance and then you center it mean=0, variance=1
Arcsine square root (rarely in linear models)
Square root of the relative abundance or raw count?
Log transformation (linear model, t-test, U, etc.)
Take the natural log and make the data look more normal. Fairly rare.
Statistical test[edit | edit source]
- Statistical test or computational tool used for differential abundance testing.
- Use the drop down options to select from. Leave blank if the study doesn’t specify.
- Some experiments (particularly multigroup experiments) can use more than one statistical test.
Significance threshold[edit | edit source]
- p-value, q-value, or FDR threshold should be reported numerically.
- LEfSe doesn’t do multiple testing correction. However, it is possible to take the p-value from LEfSe and adjust them for multiple testing in R. This is uncommon.
MHT correction[edit | edit source]
- Multiple Hypothesis Testing (MHT) correction
- Select ‘Yes’, ‘No’, or ‘Blank’
- "No" should be selected if there is no evidence of MHT correction
- It should only be left blank if the study did not use traditional statistical testing to determine significance (i.e. they used a Bayesian method or an effect size threshold instead)
- For more information on what multiple hypothesis testing correction is and various methods for MHT correction see: https://multipletesting.com/publication
Identifying MHT correction in a paper[edit | edit source]
- The use of MHT correction is usually denoted by the name of the correction in the methods or results (e.g. "Bonferroni", "False Discovery Rate," "FDR," "Benjamini-Hochberg")
- It is also present if the authors use q-values instead of p-values or mention a correction for multiple hypotheses.
Common pitfalls[edit | edit source]
- Some statistical tests such as DESeq2 correct for MHT by default while others such as LEfSe do not.
- Occasionally, some studies will report both MHT-corrected and uncorrected p-values. Generally, we curate the MHT-corrected list.
LDA score[edit | edit source]
- Threshold for the linear discriminant analysis (LDA) score for studies using the popular LEfSe tool.
- Only applies to studies using a LEfSe analysis for their statistical test.
- Numeric value only. Do not enter inequality signs (e.g. >, <, etc. are not allowed)
- For more information on LEfSe and LDA scores, see the original LEfSe paper: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3218848/
Matched On[edit | edit source]
- Matching typically only occurs in a case-control study.
- Matching occurs when they select controls similar to cases and match them on certain characteristics such as race, age, health status, gender, etc.
- It will often be phrased along the lines of “Cases were matched to controls on race, age, and gender” or “Age, gender, and race-matched study population”.
- Ctrl+F for the word “match” is generally an easy way to look for matching.
- Use the drop down to select the matched on terms. Don’t use abbreviated terms in the field.
Confounders Controlled for[edit | edit source]
Controlling for confounders can mainly occur through one of two ways:
- Stratification. They stratify on a variable (often race, age, gender, medication status) and report differential abundance results for each strata (group) in separate tables/figures/sentences.
- For instance, they report different results for men and women.
- Stratification can be done with nearly any type of analysis.
- Model adjustment. They include the confounder as a variable in a regression model.
- This will be stated in the methods and will state that they used a regression model (negative binomial, generalized linear, etc.) and this model was adjusted for a list of variables.
- That list of variables is the confounders controlled for
- You cannot adjust for confounders using bivariate tests including T-Tests, Fisher’s Exact Test, Kruskall-Wallis, Mann-Whitney, LEfSe, and chi-square.
Alpha Diversity[edit | edit source]
- Only looking at diversity in Group 1 (exposed group)
- If there is no alpha diversity test conducted, leave the sections blank.
- If an alpha diversity test was conducted with statistically significant results, put “increased” in the Group 1 (exposed group) of higher diversity, and “decreased ” for the Group 1 (exposed group) of lower diversity.
- If there is an alpha diversity test without statistically significant results between the two groups, put “unchanged” for that test.
Add a new Signature[edit | edit source]
- Add a new signature(on the experiment page, click on add a new signature).
- On the Add a new signature page, You enter the microbiome details.
Source[edit | edit source]
- The figure/table number where the signatures are found Abundance in Group 1
- Whether the abundance has increased or decreased in the Group 1 (exposed group)
- A separate signature should be created for the increased and decreased group.
- For example: Signature 1 is for increased abundance in exposed group. Signature 2 is for decreased in exposed group.
NCBI[edit | edit source]
- Curate all taxa no matter of their taxonomic levels.
- When you start typing in the name of a taxon, autocomplete will show it if if already exists in bugsigdb.org. If autocomplete does not show what you are looking for, search for it in the NCBI taxonomy browser then enter the integer NCBI taxid.
- Enter only the lowest (i.e. most specific) taxonomic rank mentioned on the paper (e.g. for genus level enter Clostridium, for species level enter Staphylococcus aureus or Lactobacillus zeae).
- You might sometimes have more than one presented comparison for the same contrasting group, so if results are largely agreeing, curate as one. if results are very different, curate separately.
- If signatures are reported using two different statistical tests and they are FULLY overlapping, record one experiment and include both statistical tests in the statistical analysis section for that experiment. Curate as two different experiments when results are different (not fully overlapping) between the tests.
Sometimes papers report an unclassified species, but provide classification up to the genus level. Such unclassified species should be excluded from the curation as opposed to curating the taxon up to the genus level. In general, differential abundance of a certain species does not necessarily imply differential abundance of the corresponding genus. (Note that this equally applies to the case of unclassified higher taxonomic levels such as the case of an unclassified genus where classification is provided up to the family level).
- If a taxon is reported as something like this Prevotella KQ959344_s, and you cannot find this in the NCBI taxonomy browser, report it exactly the way it’s reported in the paper since it could be referring to a certain strain of Prevotella.
Tips[edit | edit source]
- If two different figures/tables differ only in which taxonomic level they provide (e.g. one figure for species, one supplemental table for genera, but these resulted from the same contrast), these can be combined into a single signature.
- If taxa are not already found in bugsigdb.org, autocomplete won't show them. Any time autocomplete doesn’t work, you have to search NCBI taxonomy browser and enter the integer ID if you can find it there. It is helpful to start typing the genus and species, then cut and paste it into the NCBI taxonomy browser if autocomplete didn’t show it. The NCBI taxonomy is too large for autocomplete, so bugsigdb.org only autocompletes things already in the wiki and instead expects either integer NCBI taxids, or uninterpreted text.
- Where you see “Unknown” and a genus level, referring to an unknown species from a known genus, this is often referred to by adding “sp.” to the genus name. For example instead of “Unknown Subdoligranulum” there is “Subdoligranulum sp.” in the taxonomy browser (ID 2053618) and this ID can be entered into the bugsigdb.org signature.
- For species that are only classified to higher levels of the taxonomy than genus (for example only their family or order are known), try adding “bacterium” to the family in your NCBI taxonomy browser search, order etc. E.g. see Clostridiales bacterium (Taxonomy ID: 1898207, unclassified species from the Eubacteriales order). Eubacteriales is a heterotypic synonym for Clostridiales. Note: this should only be done when the paper presents the taxon at the species level. If the taxon in the paper is at a different rank, the curated taxon should be at that same rank.
- Sometimes the genus names were abbreviated in the signature, e.g. “F. nucleatum” instead of “Fusobacterium nucleatum”. This is supposed to be done only after the first mention of the genus/species binary name, but in this signature there was no full name elsewhere, so the easiest way was to go to the NCBI taxonomy and look up the species.
- The NCBI taxonomy names sometimes include square brackets, even if they are not part of the published signature. For example [Ruminococcus] torques. At the bottom of this NCBI page it clarifies that “Square brackets ([ ]) around a genus indicates that the name awaits appropriate action by the research community to be transferred to another genus.” This is fine, the NCBI Taxonomy ID will remain and the genus will be automatically corrected in the future just by using the NCBI taxonomy. Note that bugsigdb.org stores NCBI Taxonomy IDs, even though it instead shows current names to the user.
- Sometimes searching for a name in the NCBI taxonomy will take you instead to a synonym, which is fine. For example, try searching for “Bacteroides symbiosus”; it takes you to to the page for [Clostridium] symbiosum, which lists Bacteroides symbiosus as a “homotypic synonym”. The important thing here is that you have the correct Taxonomy ID, which refers both to the current correct name and this homotypic synonym. There are other kinds of synonyms too, Google provides good definitions (usually the first one is from wiktionary).
- Genus and higher-level taxonomic names (everything but species) are always capitalized. It can be convenient to start entering these names in bugsigdb.org uncapitalized, so that you know if it is successfully autocompleted by the introduction of capitalization. This is important because it’s possible to enter a valid name without autocomplete, that is never matched to the NCBI taxonomy in bugsigdb.org, and is instead treated as plain-text and not a part of the taxonomy.
Bulk signature entry[edit | edit source]
You can enter a signature in bulk if it is represented by comma-separated integer NCBI taxonomy IDs. Copy and paste the comma-separated list of IDs into the signature taxid form, then take care the last value is entered correctly (you may have to press space to see the integer autocomplete, or delete a space if one was pasted in).