Category talk:Pages with missing NCBI ID
Use this space to make notes about pages where you tried but were unable to resolve missing NCBI IDs from Category:Pages_with_missing_NCBI_ID. If you succeed in resolving all missing NCBI IDs in a signature, the signature will disappear from that page. See https://en.wikipedia.org/wiki/Open_nomenclature for help with nomenclature conventions, and use the NCBI Taxonomy Browser. Here is a 2-hour video of Levi working on mapping to the NCBI taxonomy.
Some useful nomenclature[edit source]
| Nomenclature | Meaning | Example | How to resolve | Link for more info |
|---|---|---|---|---|
| UCG | Uncultured genus-level group | Ruminococcaceae UCG-014 sp | This example can be treated as an uncultured Ruminococcaceae bacterium, which is listed in NCBI under a heterotypic synonym uncultured Oscillospiraceae bacterium. This is good because it has the correct rank (species) and identifies the finest-known level of taxonomy (family Oscillospiraceae / Ruminococcaceae) | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6407505/ |
| Incertae sedis or problematica | Latin for 'of uncertain placement' | I decided to call the Gemellaceae family Bacilli incertae sedis because references to it were usually under the Bacilli class but otherwise uncertain | a term used for a taxonomic group where its broader relationships are unknown or undefined. | https://en.wikipedia.org/wiki/Incertae_sedis |
| Example | Example | Example | Example | Example |
changed Ruminococcaceae UCG-014 to uncultured Oscillospiraceae bacterium[edit source]
Uncultured Ruminococcaceae bacterium is its heterotypic synonym in NCBI and as stated in the table above.--Joan Chuks (talk) 16:11, 10 March 2024 (UTC) The gut microbiota is associated with psychiatric symptom severity and treatment outcome among individuals with serious mental illness/Experiment 5/Signature 1 and The gut microbiota is associated with psychiatric symptom severity and treatment outcome among individuals with serious mental illness/Experiment 6/Signature 1--Joan Chuks (talk) 15:32, 13 March 2024 (UTC) https://bugsigdb.org/Study_961/Experiment_1/Signature_2 Changed unidentified Ruminococcacea to unclassified Oscillospiraceae. Unclassified Oscillospiraceae is a heterotypic synonym to unclassified Ruminococcaceae
Unresolved nomenclature existing in bugsigdb.org[edit source]
Signatures from the top of the list until this signature have been previously investigated but remain unsolved:
- Association Between Gut Microbiota and Helicobacter pylori-Related Gastric Lesions in a High-Risk Population of Gastric Cancer/Experiment 4/Signature 1
The following signatures have unmapped taxa that someone has looked into but been unable to resolve:
Paraprevotellaceae family[edit source]
- Diet, physical activity and screen time but not body mass index are associated with the gut microbiome of a diverse cohort of college students living in university housing: a cross-sectional study/Experiment 2/Signature 2: looks like Prevotellaceae family but I'm not sure... (Levi)
- Diet, physical activity and screen time but not body mass index are associated with the gut microbiome of a diverse cohort of college students living in university housing: a cross-sectional study/Experiment 2/Signature 2: "f_paraprevotellaceae" was changed to "unclassified paraprevotella". "unclassified paraprevotella" has no rank and In greengenes taxonomy, every genus associated with paraprevotellaceae is a candidate genus with no formal name.(Afuape Esther)
- Gut microbiota-associated taurine metabolism dysregulation in a mouse model of Parkinson's disease: a laboratory experiment study/Experiment 2/Signature 2: Paraprevotellaceae was changed to unclassified Paraprevotella. unclassified Paraprevotella has no rank. (Victoria)
The order Bacteroidales, which is part of the phylum Bacteroidetes, is home to the Paraprevotellaceae family of bacteria. These bacteria are widespread in the human gut microbiota and aid in the fermentation of complex carbohydrates like dietary fiber. They are anaerobic microbes, which means they flourish in oxygen-free environments.
Among the bacteria in the family Paraprevotellaceae are the genera Paraprevotella, Alloprevotella, and Ezakiella. These bacteria are currently being studied, and their particular roles and functions within the gut microbiome are unknown......Adanwa
Mogibacteriaceae family[edit source]
- The gut microbiota in conventional and serrated precursors of colorectal cancer/Experiment 5/Signature 1: Mogibacteriaceae family is a new and not yet validly published nomenclature: https://lpsn.dsmz.de/family/mogibacteriaceae
- Mucosal microbiome dysbiosis in gastric carcinogenesis/Experiment 1/Signature 1
- Mucosal microbiome dysbiosis in gastric carcinogenesis
- Daily Consumption of Orange Juice from Citrus sinensis L. Osbeck cv. Cara Cara and cv. Bahia Differently Affects Gut Microbiota Profiling as Unveiled by an Integrated Meta-Omics Approach
- Air pollution during the winter period and respiratory tract microbial imbalance in a healthy young population in Northeastern China
The Mogibacteriaceae family is a phylum Bacteroidetes family of bacteria. These bacteria are found in the human microbiome, namely in the oral microbiota. Mogibacterium is one of several genera of the Mogibacteriaceae family, with Mogibacterium being the most well-known.
Mogibacterium and related bacteria can be discovered in a variety of human oral locales, including dental plaques, saliva, and the oral mucosa. They are anaerobic bacteria, which means they flourish in oxygen-free settings.
The Mogibacteriaceae family is currently being studied, and its specific roles and functions in the oral microbiome are still being investigated. The oral microbiota is complex and diverse, with various interactions and consequences on oral health and disorders such as dental caries and periodontal disease.
Anaerovoracaceae is shown in LPSN: as a heterotypic synonym for Mogibacteriaceae family (Chuvochina et al. 2024) and validly published under the ICNP.
couldn't find order gemellales[edit source]
- Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients/Experiment 1/Signature 2
- The Perturbation of Infant Gut Microbiota Caused by Cesarean Delivery Is Partially Restored by Exclusive Breastfeeding/Experiment 1/Signature 1
- Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients/Experiment 1/Signature 2
- The Perturbation of Infant Gut Microbiota Caused by Cesarean Delivery Is Partially Restored by Exclusive Breastfeeding/Experiment 1/Signature 1
- Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota/Experiment 1/Signature 2
- [https://bugsigdb.org/Study_255/Experiment_3/Signature_1 Relationship between the Cervical Microbiome, HIV Status, and Precancerous Lesions/Experiment 3/Signature 1
couldn't find ASV_209 (Clostridia), ASV_157 (Absconditabacteria), ASV_205 (Clostridia), ASV-62 (Lachnospiraceae), (F0058 (Paludibacteria)[edit source]
- Sputum bacterial load and bacterial composition correlate with lung function and are altered by long-term azithromycin treatment in children with HIV-associated chronic lung disease/Experiment 1/Signature 1
- Sputum bacterial load and bacterial composition correlate with lung function and are altered by long-term azithromycin treatment in children with HIV-associated chronic lung disease/Experiment 2/Signature 1
- [https://bugsigdb.org/Study_822/Experiment_3/Signature_1 Sputum bacterial load and bacterial composition correlate with lung function and are altered by long-term azithromycin treatment in children with HIV-associated chronic lung disease/Experiment 3/Signature 1....(Chinelsy)
"SMB53" family in order Clostridiaceae[edit source]
- A taxonomic signature of obesity in a large study of American adults multiple signatures
- Association of dietary fibre intake and gut microbiota in adults
family Dehalobacteriaceae[edit source]
- A taxonomic signature of obesity in a large study of American adults/Experiment 1/Signature 2 multiple signatures
- https://bugsigdb.org/Study_68/Experiment_3/Signature_2
family Peptostreptococcales_Tissierellales[edit source]
- [https://bugsigdb.org/Study_1117/Experiment_1 Role of the Gut Microbiota in the Increased Infant Body Mass Index Induced by Gestational Diabetes Mellitus/Experiment 1
- https://bugsigdb.org/Study_1117/Experiment_1/Signature_2 (Rahila)
genus Burkholderia_Caballeronia_Paraburkholderia[edit source]
- [https://bugsigdb.org/Study_1117/Experiment_1 Role of the Gut Microbiota in the Increased Infant Body Mass Index Induced by Gestational Diabetes Mellitus/Experiment 1
- https://bugsigdb.org/Study_1117/Experiment_1/Signature_2 (Rahila)
Findings from List of Prokaryotic names with Standing in Nomenclature (LPSN) shows the correct name for family Dehalobacteriaceae to be Peptococcaceae Rogosa 1971 (Approved Lists 1980)--Joan Chuks (talk) 14:46, 24 April 2024 (UTC)
Clostridia clusters IV, XIVa, XIVb and XVIII[edit source]
- Altered fecal microbiota composition associated with food allergy in infants
- Altered Gut Microbiota Composition Associated with Eczema in Infants/Experiment 1/Signature 1
- Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic diseaseappears/Experiment 2/Signature 1
=== Clostridium IV and Clostridium XIVb
- [1] Intestinal microbiota in patients with chronic hepatitis C with and without cirrhosis compared with healthy controls/Experiment 1/Signature 2
A google search shows that Clostridium cluster IV is also called the "clostridium leptum group" containing different species. However, the cluster does not have a NCBI ID representing the entire group on NCBI. This should still be curated as "Clostridium IV"...(Esther)
Clostridium XIVa is also called "Clostridium coccoides group". This is not yet resolved on NCBI either and should still be curated as "Clostridium XIVa"... (Esther)
Clostridium XIVb remains unresolved...(Esther Afuape)
"Clostridia group xiv, Clostridium_XlVb, Clostridium_IV, Clostridium_XlVa "[edit source]
- Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis/Experiment 1/Signature 1:There are still many Clostridium species that require re-classification. Perhaps, the genome-based method can resolve this problem in the near future....(Chinelsy)
- Tumour-associated and non-tumour-associated microbiota in colorectal cancer/Experiment 1/Signature 2 (chinelsy)
"RsaHF231" "SHA-109"[edit source]
- Ambient temperature alters body size and gut microbiota of Xenopus tropicalis/Experiment 1/Signature 2: looks like Pseudomonadota phylum (RsaHF231, SHA-109) As for ("SHA-109" ) more research shows this:Candidate phylum UBP9/SHA-109 Parks et al. 2017 but I'm not sure... (Chinelsy)
family ruminococcaceae[edit source]
- Association Between Gut Microbiota and Helicobacter pylori-Related Gastric Lesions in a High-Risk Population of Gastric Cancer/Experiment 4/Signature 1 f__Ruminococcaceae__UCG-013 is not found as a bacteria in the paper. instead, f__Ruminococcaceae__UCG-02 is on the paper which turns out to be similar to Ruminococcaceae sp. B_A14
Experiment_1/Signature_1[edit source]
Lachnospiraceae_NK4A136 was updated to Lachnospiraceae bacterium NK4A136
uncultured Erysipelotrichaceae bacterium[edit source]
Gut microbiome signatures reflect different subtypes of irritable bowel syndrome Erysipelotrichaceae_ucg_003 was updated to uncultured Erysipelotrichaceae bacterium (taxID, 331630) in experiments 1,2 and 3 (ChiomaOnyido)
Composition and metabolism of fecal microbiota from normal and overweight children are differentially affected by melibiose, raffinose and raffinose-derived fructans/Experiment 3/Signature 2 Erysipelotrichaceae UCG 003 was updated to Uncultured Erysipelotrichaceae bacterium (NCBI ID -331630)--Joan Chuks (talk) 16:44, 10 March 2024 (UTC)
Uncultured genus DSSF69[edit source]
From google searches, this appears to be an uncultured genus within the Sphingomonadaceae family, in class alphaproteobacteria...(Esther Afuape)
Lachnoclostridium lactaris[edit source]
Seems like Ruminococcus lactaris but I'm not sure as the latter does not belong to genus Lachnoclostridium. I replaced it with Uncultured Lachnoclostridium sp. with taxID 1586779 which is a good fit as it has the correct rank of species...(Esther Afuape )
Possible taxonomy lineage of TG5[edit source]
Kingdom -Bacteria; Phylum - Synergistota; class - Synergistia; Order -Synergistales, Family-Dethiosulfovibrionaceae Source: Pub med central article--Joan Chuks (talk) 01:36, 2 April 2024 (UTC)
Lachnoclostridium faecis[edit source]
Lachnoclostridium faecis was replaced with Lachnoclostridium sp. (Taxonomy ID: 2028282). This is a likely good fit as it has the correct rank of species and falls under the genus "unclassified Lachnoclostridium"--Joan Chuks (talk) 12:11, 2 April 2024 (UTC)
Marinicellaceae Family[edit source]
The Marinicellaceae family belongs to the phylum Pseudomonadota and class Gammaproteobacteria...(Victoria)
The long-term gut bacterial signature of a wild primate is associated with a timing effect of pre- and postnatal maternal glucocorticoid levels[edit source]
https://bugsigdb.org/Talk:Study_911 contains record of https://bugsigdb.org/Study_911 unresolved Taxa --Joan Chuks (talk) 07:56, 27 March 2024 (UTC)
tricky but solved[edit source]
Changed Bacillus (Priestia) megaterium to Priestia megaterium Synonyms Bacillus megaterium[edit source]
This was tricky but I think it's right.
- Microbial communities associated with honey bees in Brazil and in the United States/Experiment 1/Signature 1
Changed Bacillus (Priestia) aryabhattai to Priestia aryabhattai Synonyms Bacillus aryabhattai[edit source]
This was tricky but I think it's right.
- Microbial communities associated with honey bees in Brazil and in the United States/Experiment 1/Signature 1
Changed Acetobacterales order to Rhodospirillales order[edit source]
This was tricky but I think it's right.
Changed Pseudopropionibacterium to Arachnia[edit source]
- e.g. https://bugsigdb.org/Study_799/Experiment_1/Signature_1, gave it Taxonomy ID: 2801844 since Pseudopropionibacterium is heterotypic synonym to Arachnia
- https://bugsigdb.org/Study_799/Experiment_2/Signature_2
..(Chinelsy)
Changed unknown proteobacterium to unidentified proteobacterium[edit source]
- e.g. https://bugsigdb.org/Study_799/Experiment_1/Signature_1, gave it Taxonomy ID: 2722 since unknown proteobacterium is heterotypic synonym to unidentified proteobacterium..(Chinelsy)
Changed unknown bacteria to bacterium[edit source]
- e.g. https://bugsigdb.org/Study_799/Experiment_1/Signature_1, gave it Taxonomy ID: 1869227 since unknown bacteria is heterotypic synonym to bacterium..(Chinelsy)
Changed Brevibacterium massiliense to Brevibacterium ravenspurgense[edit source]
- e.g. https://bugsigdb.org/Study_812/Experiment_1/Signature_2, gave it Taxonomy ID: 479117 since Brevibacterium ravenspurgense is heterotypic synonym and also homotypic Synonym to Brevibacterium massiliense..(Chinelsy)
Changed Erysipelatoclostridium to Thomasclavelia[edit source]
- e.g. https://bugsigdb.org/Study_806/Experiment_3/Signature_1, gave it Taxonomy ID: 3025755 since Thomasclavelia is homotypic synonym to Erysipelatoclostridium..(Chinelsy)
Changed unknown Actinomycetales to uncultured actinomycete[edit source]
- e.g. https://bugsigdb.org/Study_799/Experiment_1/Signature_2, gave it Taxonomy ID: 100235 since unknown Actinomycetales is heterotypic synonym to unidentified actinomycetales ..(Chinelsy)
Changed Cyanobacteria to Cyanobacteriota[edit source]
- e.g. https://bugsigdb.org/Study_799/Experiment_2/Signature_2, gave it Taxonomy ID: 1117 since Cyanobacteria is BLAST name to current name Cyanobacteriota ..(Chinelsy)
Changed Proteobacteria to Pseudomonadota[edit source]
- e.g. https://bugsigdb.org/Study_799/Experiment_2/Signature_2, gave it Taxonomy ID: 1224 since proteobacteria is BLAST name to current name Pseudomonadota ..(Chinelsy), [2] [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari),
[3] [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila) [4] [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
https://bugsigdb.org/Study_927/Experiment_2/Signature_2, gave it Taxonomy ID: 1224 since proteobacteria is BLAST name to current name Pseudomonadota found in ( https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1224&mode=info )(omojokun)
Changed unclassified Proteobacteria to unclassified Pseudomonadota[edit source]
- e.g. https://bugsigdb.org/Study_1009/Experiment_3/Signature_1, gave it Taxonomy ID: 81684 [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
Changed Enterobacteriales to Enterobacterales[edit source]
- e.g. https://bugsigdb.org/Study_1009/Experiment_1/Signature_1,
- e.g. https://bugsigdb.org/Study_1009/Experiment_2/Signature_2,
gave it Taxonomy ID: 91347 since Enterobacterales is heterotypic synonym to Enterobacteriales, [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
Changed Prevotella stercorea to Leyella stercorea[edit source]
gave it Taxonomy ID: 363265 since Leyella stercorea is basionym to Prevotella stercorea, [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
Changed Prevotella stercorea CAG:629 to Leyella stercorea CAG:629[edit source]
gave it Taxonomy ID: 1263103 since Leyella stercorea CAG:629 is homotypic synonym to Prevotella stercorea CAG:629, [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
Changed Verrucomicrobia to Verrucomicrobiota[edit source]
- e.g. https://bugsigdb.org/Study_1009/Experiment_1/Signature_1, gave it Taxonomy ID: 74201 since Verrucomicrobiota is equivalent to Verrucomicrobia, [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
Changed Clostridiales to Eubacteriales[edit source]
- e.g. https://bugsigdb.org/Study_932/Experiment_2/Signature_1, [e.g. https://bugsigdb.org/Study_932/Experiment_7/Signature_1,] gave it Taxonomy ID: 186802 since Eubacteriales is heterotypic synonym to Clostridiales Prevot 1953 (Approved Lists 1980) [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari)
e.g https://bugsigdb.org/Study_956/Experiment_1/Signature_1 the clostridiales should be changed to Eubacterium nodatum (Clostridiales) according to this website https://www.sciencedirect.com/topics/medicine-and-dentistry/clostridiales. (0mojokun)
Changed Lactobacillus rhamnosus to Lacticaseibacillus rhamnosus[edit source]
- e.g. https://bugsigdb.org/Study_932/Experiment_7/Signature_1, gave it Taxonomy ID: 47715 since Lacticaseibacillus rhamnosus is homotypic synonym to Lactobacillus rhamnosus [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari)
Changed Dialister sp. Marseille-P5638 to Dialister massiliensis[edit source]
- e.g. https://bugsigdb.org/Study_932/Experiment_1/Signature_2, gave it Taxonomy ID: 2161821 since Dialister massiliensis is current name of Dialister sp. Marseille-P5638 [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari)
Changed Actinobacteria to Actinomycetota[edit source]
- e.g. https://bugsigdb.org/Study_932/Experiment_6/Signature_1, gave it Taxonomy ID: 201174 since Actinomycetota is equivalent of Actinobacteria [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari),[5][Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
Changed Lactobacillus fermentum to Limosilactobacillus fermentum[edit source]
- e.g. https://bugsigdb.org/Study_932/Experiment_6/Signature_1, gave it Taxonomy ID: 1613 since Limosilactobacillus fermentum is the current name of Lactobacillus fermentum [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari)
Changed Corynebacteriales to Mycobacteriales[edit source]
- e.g. https://bugsigdb.org/Study_932/Experiment_6/Signature_1, gave it Taxonomy ID: 85007 since Mycobacteriales is the homotypic synonym of Corynebacteriales [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari)
Changed Bifidobacterium kashiwanohense to Bifidobacterium catenulatum subsp. kashiwanohense[edit source]
- e.g. https://bugsigdb.org/Study_932/Experiment_3/Signature_2, [ https://bugsigdb.org/Study_932/Experiment_6/Signature_1,] gave it Taxonomy ID: 630129 since Bifidobacterium catenulatum subsp. kashiwanohense is basionym Bifidobacterium kashiwanohense [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari)
Changed Catabacter to Christensenella[edit source]
- e.g. https://bugsigdb.org/Study_1027/Experiment_1/Signature_1, gave it Taxonomy ID: 990721 since Christensenella is homotypic synonym to Catabacter [Correlation of gut microbiota with leukopenia after chemotherapy in patients with colorectal cancer]..(Ayibatari)
Changed unclassified Ruminococcaceae to unclassified Oscillospiraceae[edit source]
- e.g. https://bugsigdb.org/Study_1027/Experiment_2/Signature_2, gave it Taxonomy ID: 473772 [Correlation of gut microbiota with leukopenia after chemotherapy in patients with colorectal cancer]..(Ayibatari)
changed TM7a to unclassified Candidatus Saccharibacteria[edit source]
Gave it Taxonomy ID: 1895827. The genus was reported as TM7a. On researching, I found that "unclassified Candidatus Saccharibacteria" has Rank: no rank.
- e.g. https://bugsigdb.org/Study_1027/Experiment_2/Signature_1, [Correlation of gut microbiota with leukopenia after chemotherapy in patients with colorectal cancer]..(Ayibatari)
Changed Synergistetes to Synergistota[edit source]
- e.g. https://bugsigdb.org/Study_932/Experiment_4/Signature_2, gave it Taxonomy ID: 508458 since Synergistota is homotypic synonym to Synergistetes [Longitudinal and Comparative Analysis of Gut Microbiota of Tunisian Newborns According to Delivery Mode]..(Ayibatari)
Changed Lactobacillus mucosae to Limosilactobacillus mucosae[edit source]
- e.g. https://bugsigdb.org/Study_884/Experiment_2/Signature_1, gave it Taxonomy ID: 97478 since Limosilactobacillus mucosae is basionym to Lactobacillus mucosae [Alterations of the Gut Microbiota in Patients with Diabetic Nephropathy]..(Rahila)
=== Changed Bacteroides sp. NSJ-2 to Bacteroides hominis (ex Liu et al. 2022)
- e.g. https://bugsigdb.org/Study_884/Experiment_2/Signature_1, gave it Taxonomy ID: 2763023 since Bacteroides homini includes Bacteroides sp. NSJ-2 [Alterations of the Gut Microbiota in Patients with Diabetic Nephropathy]..(Rahila)
Changed Prevotella timonensis to Hoylesella timonensis[edit source]
- e.g. https://bugsigdb.org/Study_810/Experiment_1/Signature_1, gave it Taxonomy ID: 386414 since Prevotella timonensis is heterotypic synonym to Hoylesella timonensis
- e.g. https://bugsigdb.org/Study_812/Experiment_1/Signature_2,
- e.g. https://bugsigdb.org/Study_812/Experiment_2/Signature_6, ..(Chinelsy)
Changed Actinomyces turicensis to Schaalia turicensis[edit source]
- e.g. https://bugsigdb.org/Study_812/Experiment_1/Signature_2, gave it Taxonomy ID: 131111 since Schaalia turicensis is basionym to Actinomyces turicensis...(Chinelsy)
Changed Propionicibacterium lymphophilum to Propionimicrobium lymphophilum[edit source]
- e.g. https://bugsigdb.org/Study_812/Experiment_2/Signature_6, gave it Taxonomy ID: 33012, since Propionimicrobium lymphophilum is homotypic to Propionicibacterium lymphophilum ...(Chinelsy)
[edit source]
- e.g. https://bugsigdb.org/Study_812/Experiment_2/Signature_6, gave it Taxonomy ID: 137730, since Falseniella ignava is basionym to Facklamia ignava ...(Chinelsy)
Changed Actinomyces neuii to Winkia neuii[edit source]
- e.g. https://bugsigdb.org/Study_812/Experiment_2/Signature_6, gave it Taxonomy ID: 33007, since Winkia neuii is basionym to Actinomyces neuii ...(Chinelsy)
Changed Atopobium vaginae to Fannyhessea vaginae[edit source]
- e.g. https://bugsigdb.org/Study_812/Experiment_2/Signature_6, gave it Taxonomy ID: 82135 since Fannyhessea vaginae is basionym to AAtopobium vaginae...(Chinelsy)
Changed Lactobacillus vaginalis to Limosilactobacillus vaginalis[edit source]
- e.g. https://bugsigdb.org/Study_812/Experiment_2/Signature_1, gave it Taxonomy ID: 1633 since Limosilactobacillus vaginalis is basionym to Lactobacillus vaginalis...(Chinelsy)
Changed Campylobacteria to Epsilonproteobacteria[edit source]
- e.g. https://bugsigdb.org/Study_859/Experiment_5/Signature_2, gave it Taxonomy ID:3031852 since Epsilonproteobacteria is homotypic synonym:to Campylobacteria..(Chinelsy)
Changed Enterobacteriales to Enterobacterales[edit source]
- e.g. https://bugsigdb.org/Study_859/Experiment_4/Signature_1, gave it Taxonomy ID:91347 since Enterobacterales is homotypic synonym: Enterobacteriales..(Chinelsy)
"candidatus" genus[edit source]
- e.g. https://bugsigdb.org/Study_98/Experiment_1/Signature_2, removed g__candidatus. "Candidatus" is a describer and cannot be a name unto itself. I just removed it from this signature (Levi)
Changed Leuconostocaceae to Lactobacillaceae[edit source]
- e.g. https://bugsigdb.org/Study_809/Experiment_2/Signature_1, gave it Taxonomy ID:33958 since Lactobacillaceae is heterotypic synonym to Leuconostocaceae
- e.g. https://bugsigdb.org/Study_809/Experiment_3/Signature_1,
..(Chinelsy)
Changed Eubacterium_brachy_group to [Eubacterium] brachy[edit source]
- e.g. https://bugsigdb.org/Study_809/Experiment_2/Signature_1, gave it Taxonomy ID:35517 since Eubacterium brachy is current name to Eubacterium_brachy_group
- e.g. https://bugsigdb.org/Study_809/Experiment_3/Signature_1,
..(Chinelsy)
“MITOCHONDRIA” cellular organelle[edit source]
- e.g. https://bugsigdb.org/Study_509/Experiment_2/Signature_2, removed “mitochondria” which was described as an organism (bacteria) and cannot be rather They are often referred to as the "powerhouses of the cell" because they are responsible for producing adenosine triphosphate (ATP), the primary energy currency of the cell, through a process called oxidative phosphorylation. I just removed it from this signature …(Chinelsy)
"mitochondria" was not curated because it is a cell organelle and not a bacteria...(Victoria)
"CHLOROPLAST" An Organelle[edit source]
- e.g. https://bugsigdb.org/Study_835/Experiment_6/Signature_1, removed "chloroplast" which is an organelle found in the cells of photosynthetic organisms, such as plants, algae, and some types of bacteria. Chloroplasts are responsible for the process of photosynthesis, which allows these organisms to convert light energy into chemical energy in the form of glucose or other organic compounds. I just removed it from this signature …(Chinelsy)
CHLOROPLAST[edit source]
During one of the discussions in the BugSigDB community, one of the curators mapped Chloroplast to cyanobacteriota, which was confirmed by the mentors. Source: https://en.wikipedia.org/wiki/Chloroplast I agree that chloroplast should be curated as "Cyanobacteriota" and not removed... (Chioma Onyido)
"OD1"[edit source]
- Replaced this with "Parcubacteria" (NCBI:txid221216) in https://bugsigdb.org/Study_793/Experiment_1/Signature_2
https://bugsigdb.org/Study_793/Experiment_1/Signature_1
Research shows that Candidate phylum OD1 bacteria (also referred to as Parcubacteria) have been identified in a broad range of anoxic environments through community survey analysis...(Chinelsy)
unclassified Eubacteriales[edit source]
- Replaced this with "unclassified Clostridiales " (taxid 39779 ) in https://bugsigdb.org/Study_454/Experiment_1/Signature_2..(Chinelsy)
k__Bacteria;p__Tenericutes;c__Mollicutes;o__RF39[edit source]
- Replaced this with "unclassified Mollicutes" (taxid 1609545) in https://bugsigdb.org/Study_219
k__Bacteria;p__Bacteroidetes;c__Bacteroidia;o__Bacteroidales;f__S24.7[edit source]
- Replaced this with Muribaculaceae (taxid 2005473, common name "family S24-7") in https://bugsigdb.org/Study_219
Escherichia/Shigella sp.[edit source]
- https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?name=Escherichia%2FShigella+sp. (Taxonomy ID: 1940338)
- e.g. https://bugsigdb.org/Study_1009/Experiment_1/Signature_1, [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
- e.g. https://bugsigdb.org/Study_1009/Experiment_2/Signature_2, [Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome]..(Rahila)
- e.g. https://bugsigdb.org/Study_1027/Experiment_2/Signature_2, [Correlation of gut microbiota with leukopenia after chemotherapy in patients with colorectal cancer]..(Ayibatari)
- The gut microbiota and metabolite profiles are altered in patients with spinal cord injury/Experiment 1/Signature 2 (Ayibatari)
Gemellaceae Family[edit source]
Gemellaceae = 539738
Some were previously called Bacilli incertae sedis (NCBI ID: 112452) - would be great to fix now the ones of these that were originally Gemellaceae to 539738
- Gut microbiome alterations in Alzheimer's disease/Experiment 1/Signature 1
- The tongue microbiome in healthy subjects and patients with intra-oral halitosis/Experiment 1/Signature 2
- Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid/Experiment 2/Signature 1
- A taxonomic signature of obesity in a large study of American adults/Experiment 1/Signature 1
- Relationship between the Cervical Microbiome, HIV Status, and Precancerous Lesions/Experiment 3/Signature 1
- Gut microbiota in patients with Parkinson's disease in southern China/Experimet 1/ Signature 2
Clostridia XI[edit source]
- Replaced this with Peptostreptococcaceae(taxid 186804, heterotypic synonym "Clostridium cluster XI") in https://bugsigdb.org/Study_761/Experiment_4/Signature_1 (Chioma Onyido)
Replaced "UNKNOWN" TAXA with "UNCLASSIFIED" TAXIDs[edit source]
- Replaced "unknown saccharimonadia", "unknown saccharimonadales", "unknown clostridia UCG", "unknown veillonellaceae", "unknown prevotellaceae", "unknown prevotella", "erysepylobacte", "unknown bacteroidales", "unknown bacteroidia", "unknown gracilibacteria", "unknown gammaproteobacteria" and "unknown lachnospiraceae"
with "unclassified candidatus saccharimonadia", "unclassified candidatus saccharimondales", "uncultured clostridia bacterium", "unclassified veillonellaceae", "unclassified prevotellaceae", "unclassified prevotella", "unclassified erysipelotricaceae", "unclassified bacteroidales, "unclassified bacteroidia, "unclassified candidatus gracilibacteria", "unclassified gammaproteobacteria" and "unclassified lachnospiraceae", respectively in https://bugsigdb.org/Study_799 ... (Chioma Onyido)
Changed BD1-5 (GN02) to Candidate phylum Gracilibacteria[edit source]
originally reported as BD1-5 (GN02), I curated as "Candidate phylum Gracilibacteria" because Candidate phylum Gracilibacteria was formerly known as GN02, BD1-5 or SN-2 according to web sources https://bugsigdb.org/Study_678 ... (Chioma Onyido)
changed TM7 to Candidatus saccharibacteria[edit source]
The phylum was reported as TM7. On reasearching, I found that "Candidatus saccharibacteria" which has the correct rank of phylum, was formerly known as TM7. This was while reviewing https://bugsigdb.org/Study_756... (Afuape Esther)
TM7 in Study-300 changed to Candidatus Saccharibacteria (Taxonomy ID: 95818) “Candidatus Saccharibacteria” was proposed as the new phylum name for TM7 based on the genomic analysis, which suggested that these bacteria, with reduced genomes, consume primarily sugar compounds. This information was found in the study, Saccharibacteria (TM7) in the Human Oral Microbiome(Sage-Journals)--Joan Chuks (talk) 01:52, 2 April 2024 (UTC)
resolved WAL 1855D[edit source]
found WAL_1855D to be Sporobacterium sp. WAL 1855D with NCBI ID 507843. It has the correct rank of species. It appeared in https://bugsigdb.org/Study_776... (Afuape Esther)
WAL_1855D updated to Sporobacterium sp. WAL 1855D (tx id - 507843)in https://bugsigdb.org/Study_803 --Joan Chuks (talk) 17:19, 2 April 2024 (UTC)
resolved Bacteria_p_OD1[edit source]
found Bacteria_p_OD1 to be Candidatus Parcubacteria with NCBI ID 221216. It has the correct rank of phylum and appeared in study https://bugsigdb.org/Study_793... (Afuape Esther)
unclassified Rhodobacteriaceae[edit source]
un_Rhodobacteraceae: found as unclassified Paracoccaceae...(Esther Afuape)
TG5[edit source]
found TG5 to be unclassified Synergistales with the Tax ID 1151719. It appeared in the study https://bugsigdb.org/Study_157/Experiment_2/Signature_2 ... (Eniola Adebayo)
unclassified bacteria[edit source]
https://bugsigdb.org/Study_845/Experiment_1/Signature_2 Ruminococcaceae [G-1] bacterium should be changed to uncultured bacteria according to the this article https://www.cytena.com/wp-content/uploads/2022/07/B.SIGHT-Sequencing-Single-cell-Genomes-of-uncultivated-bacteria-in-the-human-oral-microbiome.pdf (omojokun)
changed TM7x to Candidatus Nanosynbacter lyticus[edit source]
Originally reported as TM7x, I curated as "Candidatus Nanosynbacter lyticus". This is because another name for TM7x is "Nanosynbacter lyticus" which is equivalent to "Candidatus Nanosynbacter lyticus". This appeared in https://bugsigdb.org/Study_977/Experiment_3/Signature_1. Web source here. (Scholastica Urua - 5 July, 2024)
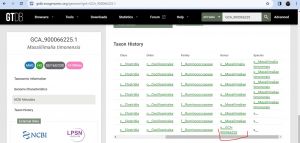
GCA-900066225[edit source]
https://bugsigdb.org/Study_844/Experiment_3/Signature_1
- I discovered GCA-900066225, as GenBank assembly accession in Genome Taxonomy Database (GTDB) for Massiliimalia. I then linked it back to NCBI,Taxonomy ID: 2895461. --Joan Chuks (talk) 16:25, 18 March 2024 (UTC)
https://bugsigdb.org/Study_999/Experiment_1/Signature_2 GCA-900066225 changed to Massiliimalia (Taxonomy ID: 2895461)--Joan Chuks (talk) 18:20, 15 April 2024 (UTC)
GCA_900066575[edit source]
I discovered GCA-900066575, as GenBank assembly accession in Genome Taxonomy Database (GTDB) for uncultured Clostridium sp. and I then linked back to NCBI,Taxonomy ID:59620.--Joan Chuks (talk) 16:14, 17 April 2024 (UTC)
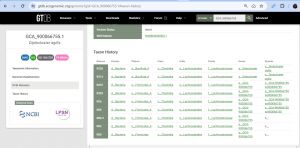
GCA_900066755[edit source]
https://bugsigdb.org/Study_774 I discovered GCA_900066755, as GenBank assembly accession in Genome Taxonomy Database (GTDB) and taxon history shows that it was a former genus name for Diplocloster, and I then linked back to NCBI,Taxonomy ID:2918511--Joan Chuks (talk) 18:47, 1 May 2024 (UTC)
Changed Eubacterium rectale ATCC 33656 to Agathobacter rectalis ATCC 33656[edit source]
- e.g. https://bugsigdb.org/Study_1131/Experiment_1/Signature_2, gave it Taxonomy ID: 515619 since Agathobacter rectalis ATCC 33656 is equivalent to Eubacterium rectale ATCC 33656
Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters ..(Rahila)
Changed Bacteroides coprocola to Phocaeicola coprocol[edit source]
- e.g. https://bugsigdb.org/Study_1131/Experiment_1/Signature_2, gave it Taxonomy ID: 310298 since Phocaeicola coprocola is basionym to Bacteroides coprocola
Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters ..(Rahila)
Changed Bacteroides coprophilus to Phocaeicola coprophilus[edit source]
- e.g. https://bugsigdb.org/Study_1131/Experiment_1/Signature_2, gave it Taxonomy ID: 387090 since Phocaeicola coprophilus is basionym to Bacteroides coprophilus
Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters ..(Rahila)
Changed Prevotella copri DSM 18205 to Segatella copri DSM 18205[edit source]
- e.g. https://bugsigdb.org/Study_1131/Experiment_1/Signature_2, gave it Taxonomy ID: 537011 since Segatella copri DSM 18205 is
equivalent to Prevotella copri CB7
Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters ..(Rahila)
Changed Eubacterium rectale to Agathobacter rectalis[edit source]
- e.g. https://bugsigdb.org/Study_1131/Experiment_2/Signature_2, gave it Taxonomy ID: 39491 since Agathobacter rectalis is
homotypic synonym to Eubacterium rectale
Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters ..(Rahila)
Changed Staphylococcales to Bacillales[edit source]
- e.g. https://bugsigdb.org/Study_1142/Experiment_1/Signature_1, gave it Taxonomy ID:1385
Differences in salivary microbiome among children with tonsillar hypertrophy and/or adenoid hypertrophy ..(Rahila)
Changed Nanoperiomorbus to Candidatus Nanoperiomorbus[edit source]
- e.g. https://bugsigdb.org/Study_1142/Experiment_1/Signature_1, gave it Taxonomy ID:2171988 since Candidatus Nanoperiomorbus is equivalent to Nanoperiomorbus
Differences in salivary microbiome among children with tonsillar hypertrophy and/or adenoid hypertrophy ..(Rahila)
Changed Nanoperiomorbaceae to Candidatus Nanoperiomorbaceae[edit source]
- e.g. https://bugsigdb.org/Study_1142/Experiment_1/Signature_1, gave it Taxonomy ID:2171987 since Candidatus Nanoperiomorbaceae is equivalent to Nanoperiomorbaceae
Differences in salivary microbiome among children with tonsillar hypertrophy and/or adenoid hypertrophy ..(Rahila)
Changed Prevotella salivae to Segatella salivae[edit source]
- e.g. https://bugsigdb.org/Study_1142/Experiment_1/Signature_2, gave it Taxonomy ID:228604 since Segatella salivae is basionym to Prevotella salivae
Differences in salivary microbiome among children with tonsillar hypertrophy and/or adenoid hypertrophy ..(Rahila)
Changed Prevotella oulorum to Segatella oulorum[edit source]
- e.g. https://bugsigdb.org/Study_1142/Experiment_1/Signature_2, gave it Taxonomy ID:28136 since Segatella oulorum is homotypic synonym to Prevotella oulorum
Differences in salivary microbiome among children with tonsillar hypertrophy and/or adenoid hypertrophy ..(Rahila)
Changed Fusobacteria to Fusobacteriia[edit source]
- e.g. https://bugsigdb.org/Study_1170/Experiment_1/Signature_1, gave it Taxonomy ID:203490
Refer: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=203490&lvl=3&lin=f&keep=1&srchmode=1&unlock Distinctive Gut Microbiota Alteration Is Associated with Poststroke Functional Recovery: Results from a Prospective Cohort Study/Experiment 1/Signature 1 ..(Rahila)
Changed Clostridium sensu stricto 1 to Clostridium[edit source]
https://bugsigdb.org/Study_1188 Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study gave it Taxonomy ID:1485 (Rahila)
Changed Prevotella loescheii to Hoylesella loescheii[edit source]
- e.g. https://bugsigdb.org/Study_1142/Experiment_1/Signature_2, gave it Taxonomy ID:840 since Hoylesella loescheii is homotypic synonym to Prevotella loescheii
Differences in salivary microbiome among children with tonsillar hypertrophy and/or adenoid hypertrophy ..(Rahila)
Changed Prevotella salivae to Segatella salivae[edit source]
- e.g. https://bugsigdb.org/Study_1142/Experiment_2/Signature_1, gave it Taxonomy ID:228604 since Segatella salivae is basionym to Prevotella salivae
Differences in salivary microbiome among children with tonsillar hypertrophy and/or adenoid hypertrophy ..(Rahila)
Resolution attempt of Stratification of the Gut Microbiota Composition Landscape across the Alzheimer's Disease Continuum in a Turkish Cohort[edit source]
Experiment_1/Signature_1[edit source]
For Prevotella_7: the taxonomic hierarchy(Kingdom to Genus) provided in the study's supplement table was the same with that of Prevotella in NCBI website, except the phylum level. The study recorded the phylum as Bacteroidetes while NCBI recorded the phylum for Prevotella as Bacteroidota. From further research (the-scientist.com), I found out that Bacteroidota is the new name for the phylum of bacteria that was previously known as Bacteroidetes. So, I updated it as Prevotella with the NCBI ID (838).
This was similar for Escherichia/Shigella, as the research showed that Pseudomonadota (Phylum name for Escherichia/Shigella sp. in NCBI) is the new name for the phylum of bacteria that was previously known as Proteobacteria (Phylum name in the study table). So, I updated it as Escherichia/Shigella sp with the NCBI ID (1940338).
Erysipelotrichaceae_unclassified seemed more like a case of a reverse of naming convention, as it appears as unclassified Erysipelotrichaceae in NCBI (ID - 544447), and was updated accordingly.
--Joan Chuks (talk) 17:46, 24 October 2023 (UTC)
Experiment_1/Signature_2[edit source]
Stratification of the Gut Microbiota Composition Landscape across the Alzheimer's Disease Continuum in a Turkish Cohort/Experiment 1/Signature 2 Mollicutes_RF39_Unclassified was updated to unclassified Mollicutes which is the closest name to the family name recorded in the study's supplementary table S2c --Joan Chuks (talk) 15:38, 2 November 2023 (UTC)
Experiment_2/Signature_1 (Clostridia cluster)[edit source]
Experiment_2/Signature_2[edit source]
Stratification of the Gut Microbiota Composition Landscape across the Alzheimer's Disease Continuum in a Turkish Cohort/Experiment 2/Signature 2 Mollicutes_RF39_Unclassified was updated to unclassified Mollicutes which is the closest name to the family name recorded in the study's supplementary table S2d
Some signatures are still unresolved - Ruminococcaceae_UCG-013, Lachnospiraceae_UCG-010, Erysipelotrichaceae_UCG-003, GCA-900066575, Ruminococcaceae_UCG-005 3068309, --Joan Chuks (talk) 16:03, 2 November 2023 (UTC)
Ruminococcaceae_UCG-005 & Ruminococcaceae_UCG-013 now updated to Ruminococcaceae bacterium UCG-005 (NCBI ID: 3068309
I discovered GCA-90006657 in Genome Taxonomy Database (GTDB) and then linked back to NCBI ID - 59620 which corresponds with the study's taxa hierarchy for the signature.
--Joan Chuks (talk) 01:13, 7 November 2023 (UTC)
Clostridium sensu stricto 1 has been solved as Clostridium genus with NCBI ID OF 1485. Refer to [6] [7] (Idiaru Angela)
Some signatures are still unresolved - Prevotella 9, Prevotella 7 https://bugsigdb.org/Study_1188 Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study
In this study i curated we have Prevotella too. (Rahila-me)
Some signatures are still unresolved - Prevotellaceae UCG-001 is a genus in the family Prevotellaceae https://bugsigdb.org/Study_1188 Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study Refer: https://midasfieldguide.org/guide/fieldguide/genus/prevotellaceae_ucg-001 (Rahila-me)
Some signatures are still unresolved - Incertae Sedis https://bugsigdb.org/Study_1188 Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study Refer: https://www.wikidoc.org/index.php/Incertae_sedis (Rahila-me)
Some signatures are still unresolved in BugSigDB- UCG-002, UCG-003, UCG-005,Lachnospiraceae UCG-010, Lachnospiraceae AC2044 group, Lachnospiraceae UCG-001, Lachnospiraceae UCG-004, NK4A214 group, Lachnospiraceae FCS020 group, Christensenellaceae R-7 group https://bugsigdb.org/Study_1188 Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)
Some signatures are still unresolved - Lachnospiraceae UCG-008,Rikenellaceae RC9 gut group, Prevotellaceae NK3B31 group https://bugsigdb.org/Study_1072 A gut microbiome signature for HIV and metabolic dysfunction-associated steatotic liver disease (Rahila-me)
Lachnospiraceae NK4A136 group has been resolved as Lachnospiraceae bacterium NK4A136 with NCBI ID of 877420. which is the closest name. Refer to (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=877420) [8] Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)
[Ruminococcus] torques group has been resolved as [Ruminococcus] torques with NCBI ID of 33039. which is the closest name. Refer to https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=33039&lvl=3&lin=f&keep=1&srchmode=1&unlock [9] Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)
[Eubacterium] ventriosum group has been resolved as [Eubacterium] ventriosum with NCBI ID of 39496. which is the closest name. Refer to: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=39496&lvl=3&lin=f&keep=1&srchmode=1&unlock [10] Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)
[Clostridium] innocuum group has been resolved as [Clostridium] innocuum with NCBI ID of 1522. which is the closest name. Refer to: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1522&lvl=3&lin=f&keep=1&srchmode=1&unlock [11] Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)
[Eubacterium] siraeum group has been resolved as [Eubacterium] siraeum with NCBI ID of 39492. which is the closest name. Refer to:
https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1522&lvl=3&lin=f&keep=1&srchmode=1&unlock
[12]
Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study
(Rahila-me)
[Eubacterium] eligens group has been resolved as Lachnospira eligens with NCBI ID of 39485. I discovered [Eubacterium] eligens, as GenBank assembly accession in Genome Taxonomy Database (GTDB) for Lachnospira eligens. I then linked it back to NCBI Taxonomy ID: 39485. Refer to: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=39485&lvl=3&lin=f&keep=1&srchmode=1&unlock https://gtdb.ecogenomic.org/genome?gid=GCA_000434415.1 [13] Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)
[Eubacterium] hallii group has been resolved as Anaerobutyricum hallii with NCBI ID of 39488. I discovered [Eubacterium] hallii group, as GenBank assembly accession in Genome Taxonomy Database (GTDB) for Anaerobutyricum hallii. I then linked it back to NCBI Taxonomy ID: 39488. Refer to: https://gtdb.ecogenomic.org/genome?gid=GCA_000435875.1 https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=39488&lvl=3&lin=f&keep=1&srchmode=1&unlock [14] Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)
[Eubacterium] brachy group has been resolved as [Eubacterium] brachy with NCBI ID of 35517 . I discovered[Eubacterium] brachy group, as GenBank assembly accession in Genome Taxonomy Database (GTDB) for [Eubacterium] brachy. I then linked it back to NCBI Taxonomy ID:35517 . Refer to: https://gtdb.ecogenomic.org/genome?gid=GCA_000488855.1 https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=35517&lvl=3&lin=f&keep=1&srchmode=1&unlock [15] Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)
[Ruminococcus] gnavus group has been resolved as Mediterraneibacter gnavus with NCBI ID of 33038 . I discovered[Ruminococcus] gnavus group, as GenBank assembly accession in Genome Taxonomy Database (GTDB) for Mediterraneibacter gnavus. I then linked it back to NCBI Taxonomy ID:33038 . Refer to: https://gtdb.ecogenomic.org/genome?gid=GCA_000433555.1 https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=33038&lvl=3&lin=f&keep=1&srchmode=1&unlock [16] Gestational diabetes-related gut microbiome dysbiosis is not influenced by different Asian ethnicities and dietary interventions: a pilot study (Rahila-me)